Sharing causal interactions with PSICQUIC
“Knowledge shared = knowledge2.” Anonymous
Defining standards is a very important initiative across scientific disciplines, since it facilitates the accessibility and sharing of information amongst data users as well as the interoperability of software tools that produce or process the respective data, thereby increasing the quality of research findings [54]. Following this logic, after the curation of causal molecular interactions from scientific literature has been achieved, the next step is to store this information to a standard data format. One of the most detailed, community-standard formats for representing molecular interaction data is the PSI molecular interaction (MI) XML format, released by the Human Proteome Organization Proteomics Standards Initiative (HUPO-PSI) [57]. The newest version of this standard is the PSI-MI XML 3.0 [58]. A simplified format for interaction data was provided by the same organization, called the Molecular Interaction Tabular format (PSI-MITAB). PSI-MITAB has become popular amongst the scientific community since it is more user-friendly and Microsoft Excel-compatible, compared to the respective XML-based format [59]. PSI-MITAB version 2.7 in particular, encapsulated many details of interest regarding a molecular interaction in a total of 42 columns, but it did not include information about its causality. This resulted in an effort to standardize the signaling information and subsequent vocabulary terminology for causal molecular interactions, originally led by SIGNOR’s database curators [60]. The new PSI-MITAB version 2.8 (also called CausalTAB), included four new columns to incorporate additional details related to a molecular interaction’s directionality (defining the biological roles of the regulator and target, one column each), regulatory mechanism (e.g. indirect causal regulation or post-translational modification) and resulting effect (up or down regulation of the target) [7].
After having a signaling data format for molecular interactions in place, the next challenge was to find a way to share such information efficiently. The heart of the problem was the same even without the addition of causality information: a large number of molecular interaction databases exist, each one with different APIs to access the respective data. Since no single database can incorporate the totality of molecular interactions pertaining to a specific biological system of interest, users have to collect data from diverse databases by launching queries in different websites or by directly downloading the respective files, which might not always be offered in standardized formats. To ease the computational access and retrieval of molecular interaction data from various resources, a web service with a common query interface (PSICQUIC) and language (MIQL) was developed [61]. Using the PSICQUIC web service, users can now download all relevant data files from their databases of interest in different PSI-MI compliant formats, suitable for further analysis (Figure 4).
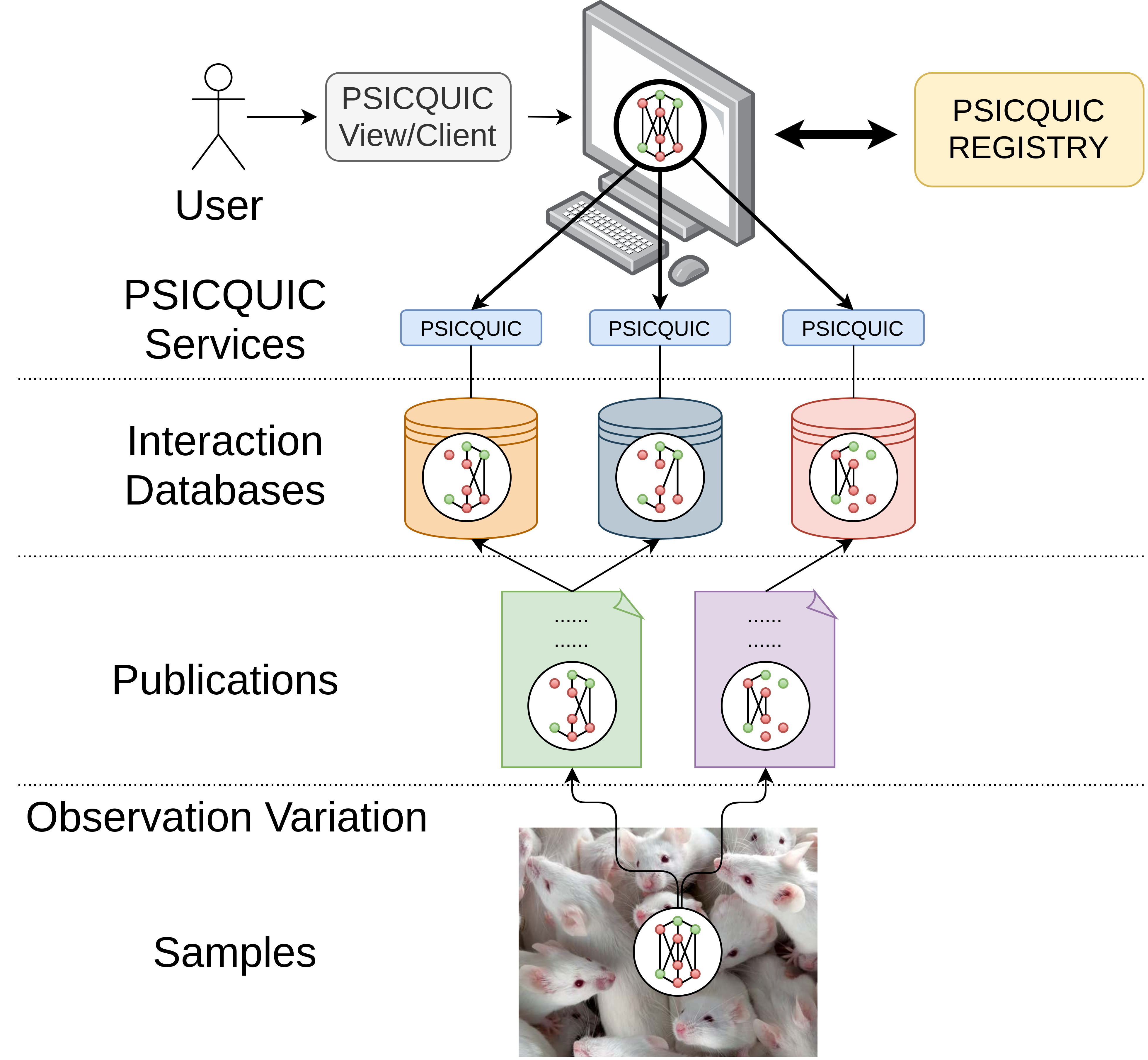
Figure 4: PSICQUIC architecture. Molecular interaction knowledge about a biological system, supported by different experimental methods, is being reported in publications. Each of these publications reports part of the actual truth about the studied system. This knowledge is curated from the respective publications and inserted to diverse molecular interaction databases. The databases share their data in standard formats (e.g. PSI-MITAB) via the PSICQUIC web service and are part of a registry list. Users launch queries via a PSICQUIC web client to retrieve the distributed molecular interaction data and synthesize the complete observed knowledge of the studied system, suitable for further analysis and visualization.
With a new signaling data format established by the relevant scientific community and the PSICQUIC web service contributing to the accessibility of molecular interaction data, the next step was to support the PSI-MITAB 2.8 format in PSICQUIC. A software project to extend the PSICQUIC platform and include causality information of molecular interactions was thus formed.4 Our contribution to this effort was the development of the underlying PSICQUIC software (version 1.4) that indexes CausalTAB files provided by the respective data providers, enabling the query and subsequent download of causality-enriched interactions. The PSICQUIC View website source code was updated to show the four new columns in the HTML table results. Additionally, several clients that enable programmatic access to the distributed signaling data (written in Java, Python and Perl programming languages), were refactored to comply with data fitting the new standard format. Lastly, the relevant PSICQUIC documentation was improved and reformatted to enhance user readability [62].
The aforementioned development effort spurred a series of actions that led to several improvements in the PSICQUIC platform. The Molecular Interactions Community, is an open source community providing tools, standard formats, ontologies and modules for manipulating molecular interaction data [63]. For example, some of these modules are used to read and write PSI-MITAB files across different versions. Since these modules had not been updated for years (showing signs of software rot [64]), we had to refactor the codebase and add tests to ensure its future reliability and quality. In the end, even though we managed to complete the task and support CausalTAB in PSICQUIC via updating these modules, the need to replace them with a newer library was imminent. JAMI is such a library, integrating all standard molecular interaction data formats such as the PSI-MI XML and PSI-MITAB, into a unified implementation, hiding the complex details of each format from the developers and thereby making their work easier [65]. The implementation work was initiated in a GREEKC workshop [66] and continued during the first ELIXIR BioHackathon in Paris [67]. Upon finishing the support for CausalTAB in JAMI,5 we provided the first PSICQUIC service indexing SIGNOR’s CausalTAB data at the time, made available through a development server [68].

During the ELIXIR BioHackathon, the architecture details of a new cloud-based, distributed PSICQUIC service were discussed and documented for future development efforts.
The goal is to enable the data providers to upload their molecular interaction data in a fully automated process and add support for data validation.
This service will minimize the long-term commitment and maintenance from the data providers, which they cannot always afford (e.g. deployment of the server hosting PSICQUIC).
Another outcome of the BioHackathon was the draft implementation of a new PSICQUIC View interface, aiming to modernize and update the current web application used to access PSICQUIC [69].
Further work needs to be done to import and use newer technologies in the interface, which will result in better filtering and sorting of the HTML table results and more interactive, graph-based visualizations of the PSICQUIC data.
To broadly facilitate the sharing of causal interaction data, additional development efforts are needed, in particular towards improving existing PSICQUIC clients.
One such example is the PSICQUIC Universal Client, a Cytoscape app for querying multiple PSICQUIC-compliant interaction data services from a simple user interface [70].
This client has been used in tutorials to guide novice users in the visualization and analysis of molecular interaction networks [71].
Lastly, two more clients that need to be updated to support the latest CausalTAB signaling format are the PSICQUIC [72] and PItools R packages [73].
These packages enable the translation of molecular interaction data directly into formats suitable for computational analysis with R, and therefore are crucial for relevant computational tasks.
This project was funded by the GREEKC (Gene Regulation Ensemble Effort for the Knowledge Commons) COST action (https://www.greekc.org/) and was realized as a Short Term Scientific Mission (STSM) in cooperation with engineers from the IntAct team at the European Bioinformatics Institute (EBI).↩︎
During the GREEKC Marseille Hackathon 2019 event, see more info on the project here: https://github.com/GREEKC/hackathon-marseille/tree/master/project_descriptions/causal_psicquic↩︎