Biological Dictionaries aid in the curation of complex knowledge
“Dictionaries are like watches; the worst is better than none, and the best cannot be expected to go quite true” Samuel Johnson
During the design process of the VSM-box tool and its application, CausalBuilder, we came across a critical technical issue that needed to be addressed, and whose resolution had ramifications outside of the intended scope of our work. Due to the high degree of complexity within the domain of biology, biocurators need to annotate diverse information, taken from a plethora of biological resources and vocabularies. To enable a wider expressiveness in the annotation of causal statements, the recommended list of ontologies and vocabularies of the MI2CAST standard had to be rather extensive [48]. Since CausalBuilder conforms to the MI2CAST standard, a unified way to retrieve, format and display vocabulary terms from different databases was needed. We illustrate this with an example: in Figure 3, a simple VSM-template that a curator can use to annotate a causal statement with CausalBuilder is shown. Following the MI2CAST guidelines, the source entity of the causal statement (first box in Figure 3) must always be specified and a list of recommended resources where the annotation could potentially originate from is provided [48]. We limit the number of these resources to three in this example, making it so that the source biological entity can be annotated as a protein (from UniProt [49]), a complex (from Complex Portal [50]), or an RNA transcript (from RNAcentral [51]). The intended use case is that the curator will type in a string (e.g. “tp53”) and a list of terms and descriptive metadata from the three respective standard databases will be returned. This information can be displayed by VSM-box in an autocomplete drop-down menu to ease the selection of the appropriate term by the curator.
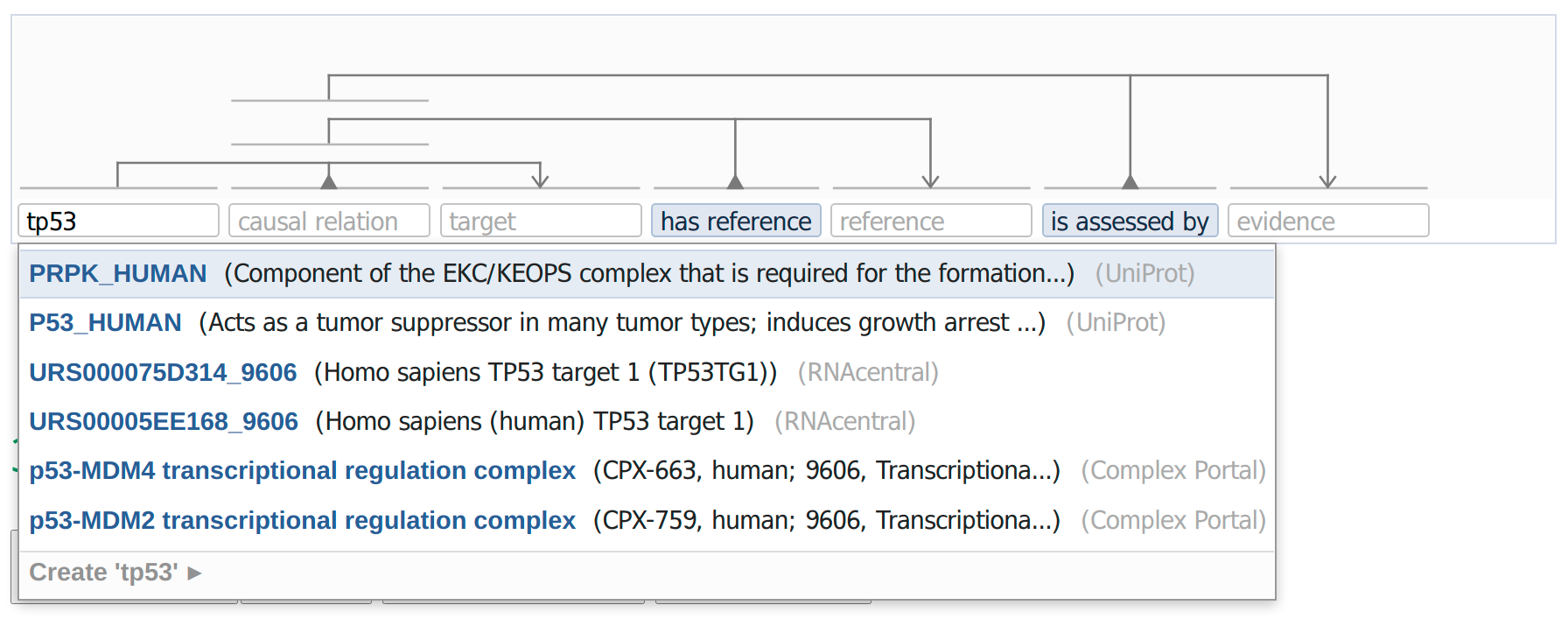
Figure 3: Querying multiple data resources using the VSM-box technology in CausalBuilder. The user enters a string of interest and selects a list of resource types (not shown here) for the source entity, following the MI2CAST curation guidelines. The UBDs stand as a hidden translator between the query launched from the curator interface and the respective database data, returning a list of uniformly-structured matches, shown as a drop-down list to the user. The matches consist of a curator-friendly main term (shown in blue) and metadata like identifier, name of species, textual description, resource name etc., that a user can use to disambiguate between the different concepts.
We can now clearly state the heart of the problem: the resources that offer protein, RNA and multiprotein complex data, have different online APIs to serve their information, and it is usually structured in diverse formats. Therefore, it was necessary to design a generic solution that would translate all the necessary information from the recommended resources of the MI2CAST standard into a unified representation schema. Then, we could implement modules that “talk” to the databases and translate the provided information into this uniform data format. As a result of having a standardized way to represent data from various disparate resources, VSM-box and other curation tools could easily process the returned data load and create drop-down menus to help users in their annotation tasks (as shown in Figure 3). The outcome of all this effort was the implementation of UBDs (Unified Biological Dictionaries, see Paper 1). The reason for the name dictionaries originates from the abstract data type called associative array (also known as map or dictionary), which is a collection of (key, value) pairs, and is an integrated feature of many programming languages. For our application, we reasoned that the minimum information that is needed for the unique identification of concepts for curation tasks is a computer-friendly ID and a human-friendly term, precisely matching the key and value of the associative array’s data structure.
An unforeseen consequence of the UBDs implementation was that by covering most of the vocabularies and ontologies recommended by the MI2CAST standard, we ended up mapping into a unified format a large amount of diverse terminologies across life sciences. This happened because our solution encapsulated and extended other similar efforts, such as the BioPortal [52] and EBI Search [53] web services. We therefore managed to bring even more biomedical ontologies and biological data resources under one umbrella, and subsequently increase the accessibility, interoperability and reusability of the provided data [54]. So, even though UBDs main user is the software engineer building curation tools (as we were at the beginning of this effort with CausalBuilder), several computational researchers can benefit from our implementation, if they need to query disparate biological resources for lightweight information (i.e. terms, identifiers and some metadata) using a single programmatic interface. In the end, the feedback we got from biocurators who tested the CausalBuilder tool was very encouraging, pointing out that we had proceeded in the right direction with our efforts to build UBDs, the hidden machinery enabling all the autocomplete “magic” to happen in VSM-box’s user interface.
The implementation of UBDs put us in a position to confront problems that biocurators face during their annotation tasks, which haven’t yet been properly addressed by any existing technology. One of these challenges is that biocurators often need to annotate terms in a specific domain or novel field, for which there is still no authoritative database or ontology nor a community consensus about the respective terminology [55]. A similar challenge manifests when new knowledge is discovered or similarly, further contextual information related to existing knowledge comes into light, as a result of scientists’ constant drive for progress. This eventually leads to the constant refactoring of ontologies and identifiers, subsequently making biocurators life even more difficult. To respond to these challenges, biocurators create project-specific, ad-hoc vocabularies that are not openly accessible and usually become obsolete after some time passes. We reasoned that with the UBDs infrastructure in place, we could do better.

In summary, the core of the problem is two-fold: first, biocurators need a simple way to annotate new information that does not yet exist in any resource and second, this information needs to be shared publicly for further review and management by expert communities. To tackle this problem, we collaborated with experts from PubDictionaries, an online repository of publicly accessible and editable dictionaries [56]. Using the online interface of PubDictionaries, curators can create simple dictionaries, consisting of terms and identifiers of their own choice, solving the second part of the problem. Additionally, by updating the PubDictionaries API and connecting all existing and future public dictionaries with UBDs and their underlying unified format, we streamlined their use in annotation tools and solved the first part of the problem. The technical work was carried out during an intense hacking week at the ELIXIR Biohackathon 2021 event and the implementation details are described in Paper 2 of this thesis. As a final result, we showcased a demo in which curators could use their public, ad-hoc terminologies from PubDictionaries, to annotate a simple sentence using the VSM-box interface.
The following video is the presentation of the demo during the last day of the ELIXIR Biohackathon 2020: