Knowledge from a stack of papers
“We know more than we can tell.” Michael Polanyi
Where does the information that is used to build knowledge networks originate from? One of the most widely adopted ways to record and share knowledge, has been the publication of scientific findings in specialized journals. This has resulted in a major challenge that researchers in the life sciences face, which is to stay updated with the huge amount of information that is published on a daily basis (Figure 2). It becomes impossible for the average scientist to find, read, extract and use that information in an efficient manner without the use of databases. Even when using databases, one is often confronted with both chronically incomplete knowledge, and also a lack of sufficient contextual information to assess when exactly the knowledge is valid.
![Human vs Life-Science Literature. How can humans stay up-to-date with increasing knowledge stored in PDF files? [36]](img/papers.png)
Figure 2: Human vs Life-Science Literature. How can humans stay up-to-date with increasing knowledge stored in PDF files? [36]
A severe problem lies already at the data entry stage. Biocurators are people whose main task is to read the scientific literature and translate knowledge into a precise, computable form, ready to be inserted into databases [37,38]. The huge body of literature existing today is full of inconsistencies and inaccuracies, so expert interpretation and annotation are essential. But current databases are limited in what they can contain, because there exists no easy way to properly transfer all kinds of complex knowledge or ideas into them, in the first place. Moreover, the annotation tools that biocurators use are not intuitive nor flexible enough to be used by large crowds of people, to convert vast amounts of relevant knowledge from the scientific literature into the respective databases. The insufficient funding to curate scientific results into databases, and the cost of creating a new knowledge base for every new project, are some extra confounding factors. Because of this, researchers all over the world have to spend considerable time performing ad-hoc manual curation of publications that are relevant for their project, often with improvised approaches (Word, Excel). At best they also spend time developing a specialized curation platform or computational methods to extract knowledge, which can only capture a fraction of the “actual reported truth” [39]. Nonetheless, all these efforts form a significant part of the scientific enterprise, assisting in the creation of digital knowledge repositories, which are subsequently used to build PKNs for the computational modeling of biological processes.
A list of tools have been created to assist biocurators in their annotation tasks. Notably, the IntAct editor is an open-source desktop application software that enables IntAct curators and members of the IMEx consortium to annotate molecular interactions [40]. Because of the lack of installation instructions and documentation, coupled with a complex interface, specialized training from senior IntAct curators is required to learn how to use this software. Nonetheless, it is one of the most used and effective tools for the job, since it has been around for a lot of years and during that time, there has always been a spirit of close collaboration between developers and curators to implement features, solve bugs and in general improve the annotation capabilities of the software. Canto is another tool that was built to support community curation in the PomBase fission yeast database [41]. It has now expanded its original purpose to support curation of other model organism databases and different molecular data types (e.g. annotation of a larger set of GO terms). Canto’s respective website provides extensive documentation and step-by-step user guidance throughout the annotation procedure [42]. A user management mechanism is incorporated in the software so as to allow proper monitoring of curation tasks and efficient communication between curators for work prioritization. In addition, two relatively new tools have been developed for the curation and visualization of molecular interaction maps: NaviCell [43] and MINERVA [44]. These tools facilitate knowledge exploration in addition to knowledge annotation, allowing for an interactive user experience (e.g. feedback via comments), enabling content sharing, supporting well known data standards (e.g. SBGN [45]) and thus allowing for data interoperability and re-use. All the aforementioned annotation tools are limited by the fact that they are not generic enough to curate any type of information, with most of them representing specialized solutions pertaining to specific annotation purposes. Most tools require extra technical configurations and software to include additional levels of contextualized details required for current and future curation efforts.
To obtain support from computational pipelines that will help us process vast amounts of knowledge and advance our understanding of processes in nature, we must be able to efficiently annotate and store information that is highly detailed and contextualized. Hereby, the knowledge’s inherent complexity should be kept manageable and understandable by humans and machines alike. In order to accommodate for a much more powerful, flexible, and reusable annotation process, an intuitive curation and knowledge formalization method was developed, called VSM (Visual Syntax Method) [46]. VSM enables scientists to capture any type of knowledge with any type of contextual information, in a way that is understandable by both humans and computers.
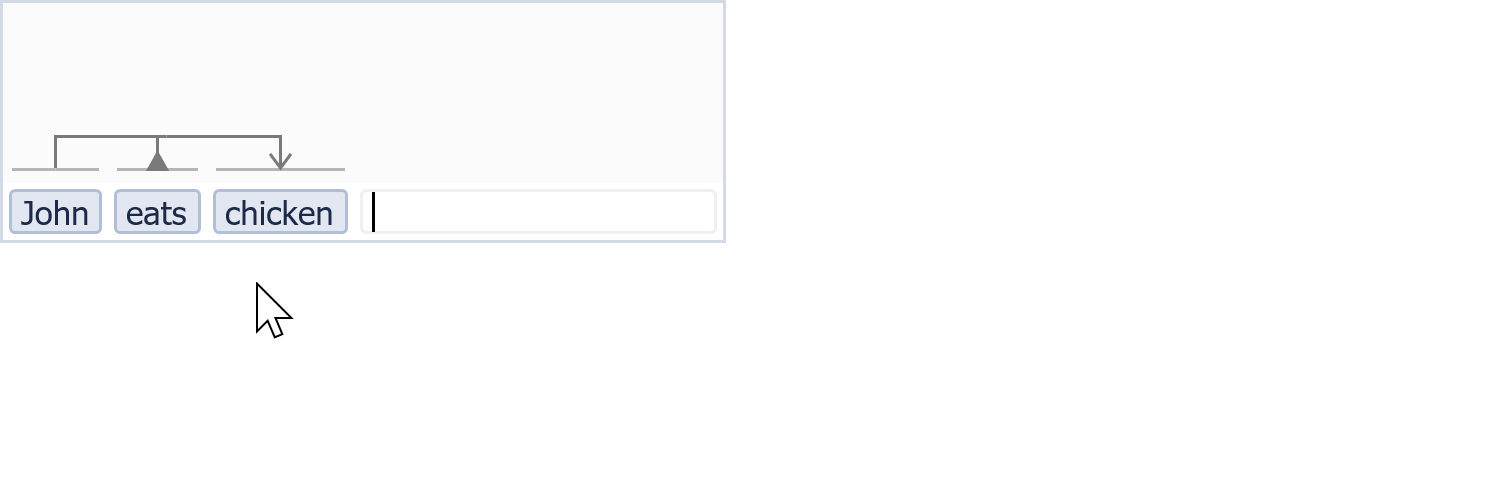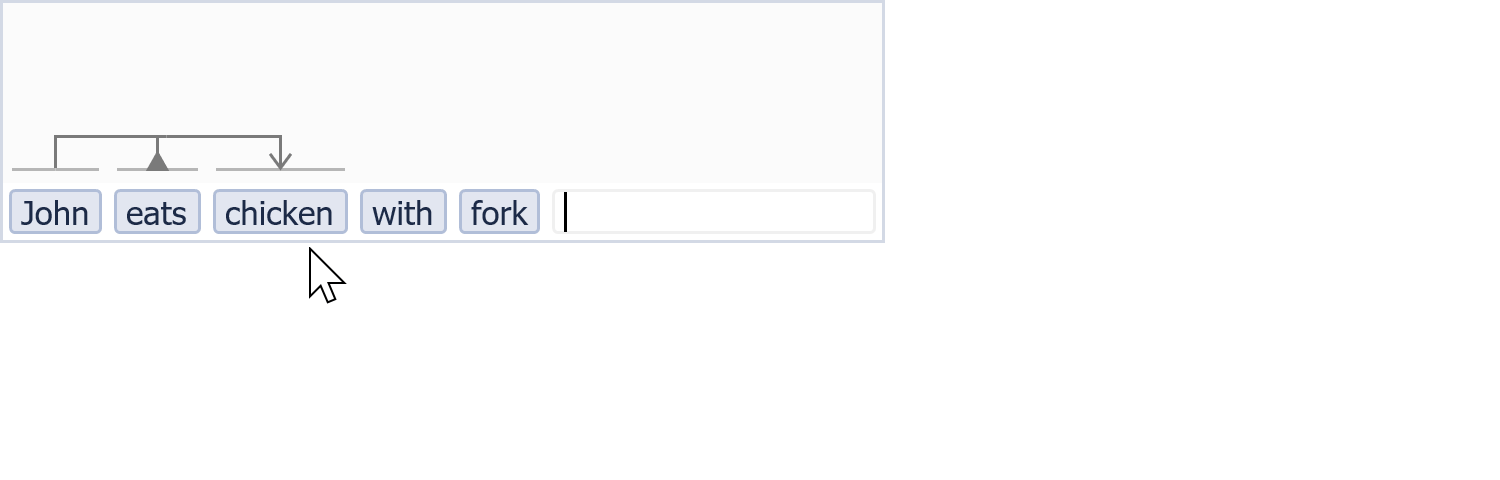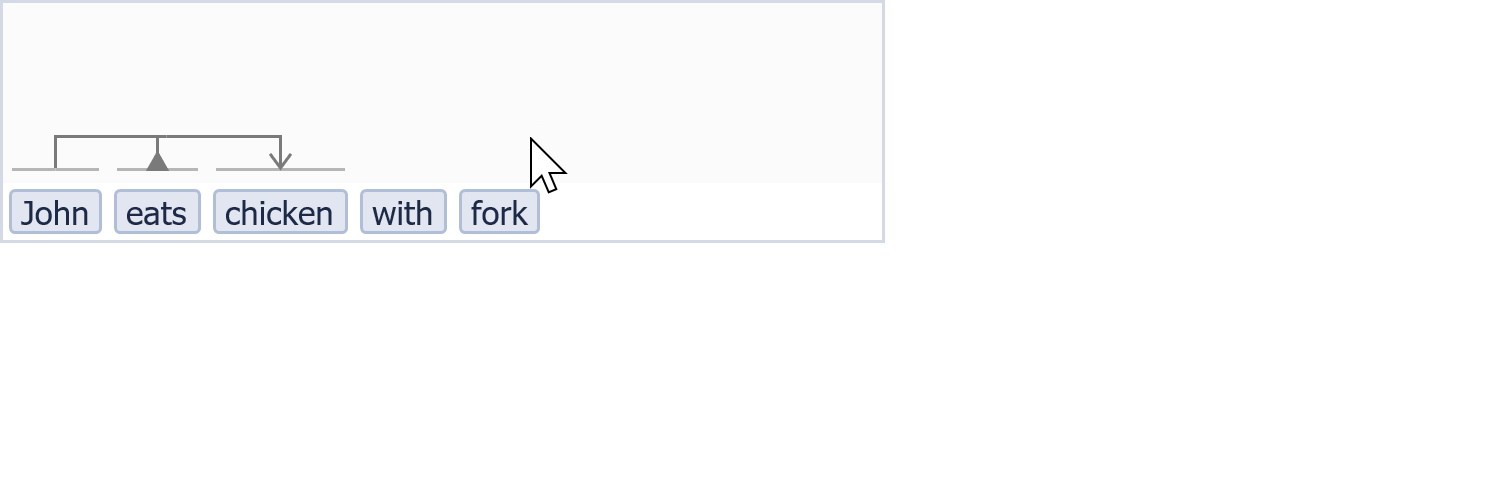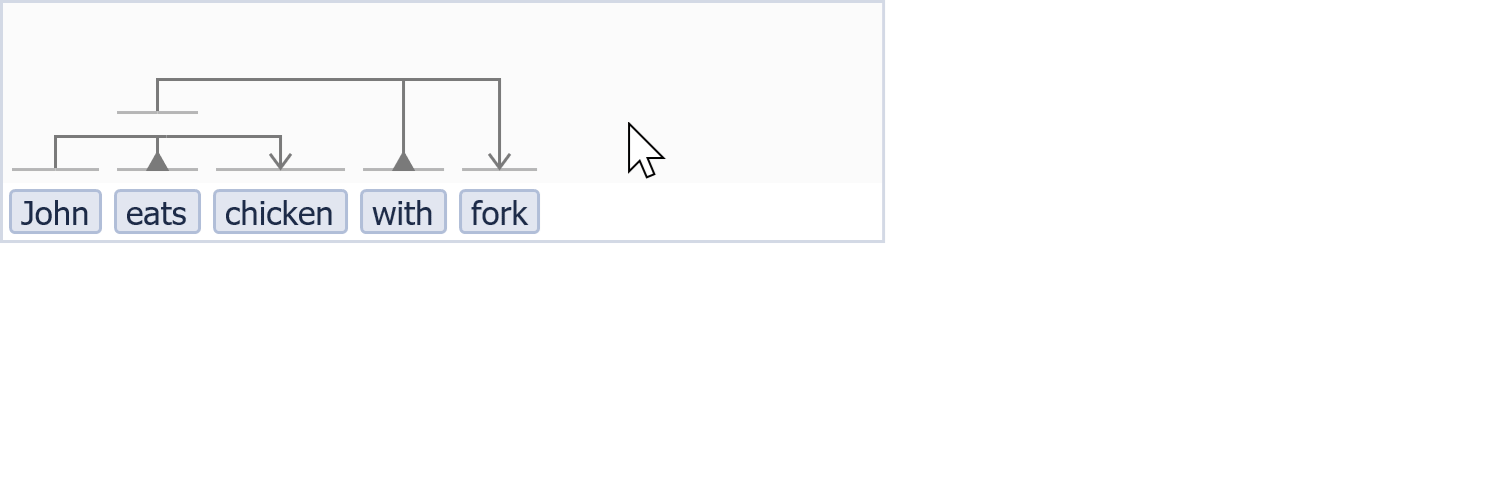
A little bit VSM in action...
Part of the work in this thesis has been to assist in the implementation of a software module that implements VSM as a general-purpose, web-based user interface, named VSM-box [5]. This software component was used to build CausalBuilder, a prototype curation interface for the annotation of causal molecular interactions [6]. CausalBuilder uses VSM to generate concrete, customizable templates that represent causal statements. It supports the export of the annotated statements in standard signaling formats, such as CausalTAB [7], which can be stored in relevant databases or used to build computational models of biological processes. To support the large variety of contextual information related to causal molecular interactions between biological entities, allowing for a finer disambiguation between seemingly similar or conflicting causality statements (e.g. a transcription factor simultaneously up and down regulating a target gene in different cellular contexts), CausalBuilder was designed to comply with a list of guidelines (MI2CAST) that were developed exactly for this purpose [47]. All in all, CausalBuilder provides biologists and curators with a simple user interface for the annotation of causal regulatory knowledge, translating highly contextual information about molecular interactions from scientific publications to a computable form.